Cells generate directed forces by regulating the alignment and motion of molecules at the expense of chemical energy. For example, epithelial cells migrate on their substrates using directed forces generated by the retrograde flow of actin filaments at the leading edge. Understanding how cells achieve anisotropic organization and orchestrate directed forces requires the ability to visualize the alignment and flow of biomolecules. Flow of molecules can be revealed using an imaging mode called fluorescence speckle microscopy that utilizes sparse labeling of molecular networks. However, imaging the molecular alignment and orientation, or ‘molecular order’ in short, in live cells remained challenging. Spatial resolution achieved by current fluorescence super-resolution methods is insufficient for quantitative analysis of molecular order in live cells. Electron microscopy can resolve molecular order, but is not compatible with live specimens.
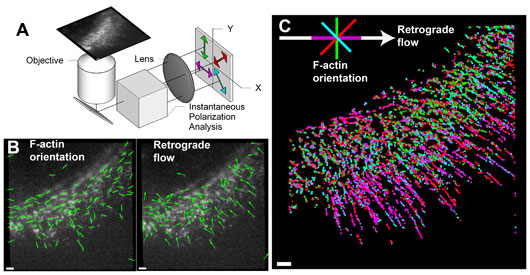
Figure: (A) Schematic of the instantaneous fluorescence polarization microscope, which simultaneously acquires four polarization-resolved views of a fluorescent specimen. (B) Retrograde flow (arrows) and orientation of actin filaments (lines) within sparsely labeled actin network at the leading edge of a migrating human skin cell. (C) Plot of the orientation of actin filaments relative to the local retrograde flow direction, colored according to the key on the top-left. The map of the actin filament orientation relative to flow direction is generated by projecting the data shown in (B) over many time-points. Scale bars correspond to 1µm.
To address this challenge, we developed a microscope that exploits intrinsic polarization of the fluorescence emission and fluorescent probes attached semi-rigidly to the molecules of interest. The microscope, termed instantaneous fluorescence polarization microscope (instantaneous FluoPolScope), acquires four polarization-resolved views of the specimen in a single snapshot (Figure A). Combined with robust algorithms for analysis of the orientation, alignment, intensity, and flow of sparsely labeled molecules, this microscope enables measurement of the molecular order within complex macromolecular assemblies (Figure B). With the new imaging method, we analyzed the orientation of actin filaments relative to the retrograde flow at the leading edge of a migrating human skin cell (Figure C). Previously, analysis of the orientation of actin filaments has only been possible in fixed cells with an electron microscope.
The instantaneous FluoPolScope is applicable to the study of emergence of order in diverse macromolecular assemblies. For example, in collaboration with Amy Gladfelter’s group (University of North Carolina, Chapel Hill), we have tracked the assembly of single septin protofilaments as they form higher-order structures in live cells [1]. In a recent multi-institution collaboration, we have used the microscope to measure the orientation of integrin transmembrane receptors when they engage with actin retrograde flow inside the cell and a rigid ligand outside the cell [2,3]. Thus, cytoskeletal polymers can ‘actively align’ protein complexes that they interact with, a phenomenon revealed only in live cells with fluorescence polarization.
Support from HFSP in the form of a Cross-Disciplinary Fellowship to the lead author, Shalin Mehta, enabled him to bring his training in optics and computation to bear upon the challenge of measuring molecular order with single molecule sensitivity. In addition, the research was supported by intramural funds from the Marine Biological Laboratory as well as National Institutes of Health’s RO1 awards to the mentors, Rudolf Oldenbourg and Tomomi Tani.
Reference
[1] Dissecting ordered organization of biomolecular assemblies in live cells by tracking position and orientation of single dipoles and their ensembles. S. B. Mehta, M. McQuilken, P. La Riviere, P. Occhipinti, A. Verma, R. Oldenbourg, A. S. Gladfelter, and T. Tani, PNAS, vol. PNAS Plus, pp. E6352–E6361, 2016.
Other References
[2] Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. *P. Nordenfelt, *T. I. Moore, *S. B. Mehta, #J. K. Mathew, #V. Swaminathan, N. Koga, T. J. Lambert, D. Baker, J. C. Waters, R. Oldenbourg, T. Tani, ^S. Mayor, ^C. M. Waterman, and ^T. A. Springer, In review, bioRxiv preprint: http://dx.doi.org/10.1101/071936, 2017.
[3] Actin retrograde flow orients and aligns ligand-engaged integrins in focal adhesions. *V. Swaminathan, *J. K. Mathew, *S. B. Mehta, #P. Nordenfelt, #T. I. Moore, N. Koga, D. Baker, R. Oldenbourg, T. Tani, ^S. Mayor, ^T. A. Springer, and ^C. M. Waterman. In review, bioRxiv preprint: http://dx.doi.org/10.1101/071852, 2017. *,#,^ indicate equal first, second, and senior authors, respectively.


































