In recent years, the possibility to mimic human organ growth in vitro in the laboratory has opened up exciting possibilities. 3D tissue structures, so-called organoids, are grown from human stem cell material. Over recent years, many protocols for modeling human organs in vitro have emerged, ranging from kidney and liver to brain and skin models. By offering scientists the possibility to perform experiments in a tissue-like context without the need for animal experiments, organoids are becoming a promising tool not only for basic biomedical science but also drug development.
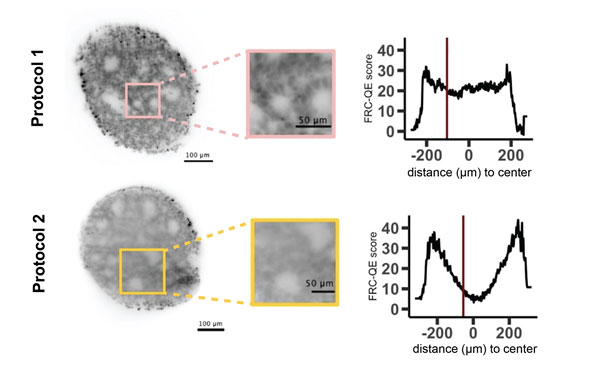
Figure: as shown in this example, different clearing protocols can give different results in terms of clearing efficiency of a given sample. FRC-QE can detect the resulting difference in image quality throughout the 3D volume of the organoid without the need for a priori knowledge about the imaged cell type or staining. The red line in the plot shows the position of the corresponding image on the left.
Since organoids have the capability to grow in vitro, organoids are well suited to three-dimensional fluorescent microscopy. High-resolution microscopes such as confocal or light-sheet based set-ups can thus provide information about the spatial distribution of proteins, enzymes or RNA molecules within a global, three-dimensional context. Nevertheless, organoids are dense and usually very opaque structures and therefore can make fluorescent imaging challenging. This challenge can be overcome by performing optical clearing, a technique that chemically treats the samples to render them translucent. Numerous sample protocols exist, from which experimentalists have to carefully select promising methods and optimize each of them for a given sample. Evaluation of these results is very time consuming and, most importantly, typically biased if performed manually.
To overcome this challenge, we developed an automated software tool for assessing image quality in cleared organoids. Our software, which is written using ImgLib2/Java and can be readily used within the popular Fiji environment, scales to large images and provides users with a robust and comparable metric for assessing and comparing the performance and repeatability of clearing protocols for a given sample, staining and microscopy set-up. We envisage this tool will be helpful for people that are about to start implementing an existing, or develop, a new clearing protocol. Additionally, we believe that the concept is applicable to other tasks where automated quality assessment of 3D image data is required.
This work was the result of a cross-disciplinary international collaboration, involving scientists from different institutions across two continents. Importantly, the project could only succeed through the combined expertise of medical doctors, biologists and computer scientists. We thank HFSP for the generous funding that allowed us to work together on this interdisciplinary endeavor.
Example Video
Tutorial
|
HFSP award information Research Grant - Program (RGP0021/2018): Quantitative dissection of molecular determinants of enhancer function Principal investigator: Nicolas Gompel, Ludwig-Maximilians-Universität München, Planegg-Martinsried, Germany (Nationality: France) |


































