When any piece of electronics such as a radio breaks, the first thing an engineer will ask for is a circuit-wiring diagram. These diagrams contain the connectivity of all components in the circuit. These diagrams are essential guides to decipher the circuit’s function and are key to devise methods to fix it. This is also true of attempts to understand our brain, whose functions are based on networks that are dramatically more complex than any electronic component ever envisioned.
The neuronal connections that enable biological circuits to function are tiny: around fifty thousand times thinner than a human hair. Thus, the resolutions required to visualize them are at present only accessible using electron microscopy (EM). This approach works as follows: brain tissue samples are treated with heavy metals to stain the membranes of neurons and their neuronal machinery. This tissue is subsequently embedded in plastic blocks that provide the stiffness required to cut very thin sections. These sections are sequentially collected on a tape, mounted on silicon wavers and imaged in an electron microscope. As the name already reveals, an electron microscope shoots beams of electrons at samples. The particular EM-technique used in these experiments detects the electrons that are scattered back after colliding with the deposited heavy metals to render an image. Finally, this imaging process is automatized to sequentially image serially cut sections, allowing the generation of volumetric datasets of brain tissues with all their components.
The approach mentioned above has been given the name ‘connectomics’, which refers to the field that studies comprehensive maps of neural connections in the brain. This approach has helped to expand our mechanistic understanding of brain computations, but with a huge cost. Even imaging and analyzing the smallest sample requires a monumental human effort, mostly because finding the relevant connections is literally as effortful as finding a needle in a haystack. The common method, called ‘saturated’ reconstruction, relies on manual or semiautomatic segmentation of all objects in such datasets, an extremely time consuming enterprise. An alternative is ’sparse’ reconstruction of specific cells or circuit motifs within a fully imaged volume. For example, neuronal activity can be monitored using calcium indicators, then neurons with particular patterns of activity can be relocated in thin sections and reconstructed. This method is, however, technically demanding and infeasible in many tissues. Thus, when multiple samples must be compared – e.g., controls vs. mutant or treated vs. untreated animals – mapping neuronal circuits remains out of reach.
To overcome this limitation we developed a new procedure named ARTEMIS that uses a combination of multiple techniques to speed up the mapping of neurons and their connections dramatically. ARTEMIS (for Assisted Reconstruction Technique for Electron Microscopic Interrogation of Structure) makes use of genetic engineering, serial-scanning electron microscopy, an enhanced chemical staining procedure and a new image processing approach. This pipeline starts by tagging a specific cell type with a genetically encoded EM tracer. These tracers are enzymes that catalyze a chemical reaction whose product can interact with heavy metals, thus making it visible in the electron microscope. We then devised a trick that enhances the electron-density of the stain without compromising ultrastructure of the surrounding tissue. This is essential to generate accurate and unbiased maps of all connections in a circuit. In addition, it allowed us to find the labeled neurons and their processes even at low resolutions. Because this is much faster than imaging them at high resolution, we could quickly generate overview maps of selectively targeted cells. These overview maps act as a road map that guides the necessary high-resolution, and therefore slower, imaging efforts onto regions encompassing the motifs of interest. Finally, we developed a novel algorithm that takes advantage of the high contrast of the targeted interneurons. It is reliable, fast and does not require computationally intense optimizations, automatizing this process even further (Fig. 1).
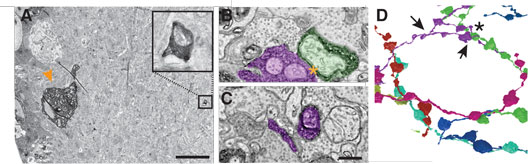
Figure 1. Finding a needle in the haystack. A. EM-micrograph of a mouse direction selective RGC soma rendered electron dense using a cytosolic EM-tag (arrowhead). Note the small process visible in the inner plexiform layer visible at this low resolution (boxes, enlarged image on the right). B & C. Two high-resolution EM-micrographs containing tagged neuronal processes (colored as in D). D. Reconstruction of the connectivity diagram with the characteristic dendritic fasciculation of the targeted retinal interneuron, called starburst amacrine cells. Asterisk indicates contact in B and arrowheads the locations of B and C. Scale bars: A: 10 μm; B-C: 200 nm.
Taken together, ARTEMIS reduces the time required to map the shapes and connectivity of neuronal motifs by two orders of magnitude, allowing comparisons across different tissues. Identifying cases where wiring goes awry leading to disease might not be a fictional endeavor anymore.
Reference
Reconstruction of genetically identified neurons imaged by serial-section electron microscopy. Joesch M, Mankus D, Yamagata M, Shahbazi A, Schalek R, Suissa-Peleg A, Meister M, Lichtman JW, Scheirer WJ, Sanes JR. Elife 5, (2016).


































