mRNA translation, the ability to translate the nucleic acid code into proteins, underlies all forms of life. The enzymes that decipher the genetic code and catalyze the first step of protein synthesis, are aminoacyl-tRNA synthetases. They recognize tRNAs and charge them with their respective amino acids, thereby providing the building blocks for the ribosome to assemble proteins following mRNA instructions. In general, each of the proteinogenic amino acids is recognized by its own aminoacyl-tRNA synthetase, bringing their number up to 20 cytoplasmic tRNA synthetases in humans. While the enzymatic core of these proteins is highly conserved, the rest of the aminoacyl-tRNA synthetase acquired new domains which enables them to perform functions beyond mRNA translation. If one traces aminoacyl-tRNA synthetases throughout evolution, it becomes clear that the increasing complexity in living beings is mirrored in their tRNA synthetases. With the appearance of new appended domains, tRNA synthetases obtained the ability to organize themselves into a multisynthetase complex. In humans, this evolutionary drive for complex formation culminates in 9 enzymatic activities being held together by 3 designated adapter proteins.
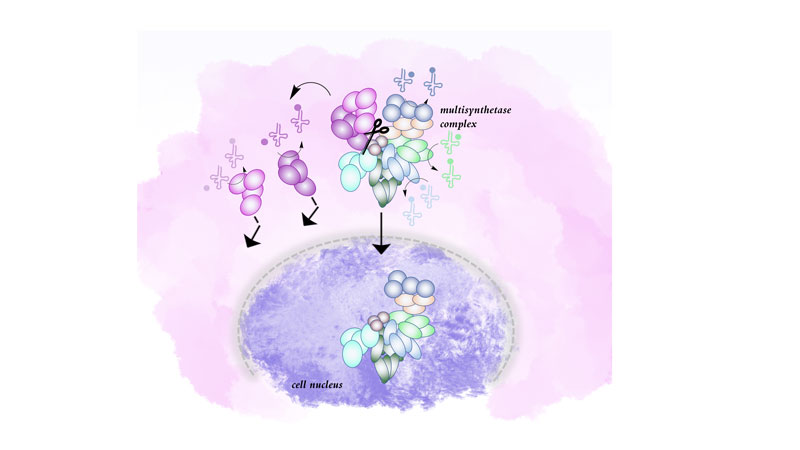
Figure: Representation of the multisynthetase complex in a human cell. Upon deletion of an anchoring domain by gene editing, the excluded proteins can no longer be integrated into the multisynthetase complex. Exclusion of aminoacyl-tRNA synthetases from the complex also leads to their exclusion from the cell nucleus where they might fulfill functions aside from tRNA charging.
It has long been assumed that this multisynthetase complex is formed to increase the efficiency by which aminoacylated tRNAs are channeled to the ribosome. We tested this hypothesis by using CRISPR/Cas9 guided gene editing to force translation from an alternative start codon in one of the tRNA synthetases in the complex, leading to the exclusive production of a shorter variant that lacks the domain that anchors the protein in the complex. In addition, another tRNA synthetase that is anchored to the complex by the “cut” enzyme is also lost but can be brought back by expression of the anchoring domain alone.
In this system, we now investigated whether the loss of one or two aminoacyl-tRNA synthetases from the complex affect 1) tRNA charging, 2) mRNA translation, focusing on the codons decoded by tRNAs cognate to the excluded tRNA synthetases, or 3) bulk protein synthesis. We found that none of these were reduced, suggesting that indeed, the absence or presence of these two tRNA synthetases from the complex had no effect on their canonical function. Therefore, we chose to interrogate non-canonical functions of aminoacyl-tRNA synthetases instead. To fulfill these, the proteins need to be in distinct locations in the cell. Using nuclear localization as a readout, we found that if aminoacyl-tRNA synthetases are excluded from the complex, they are also excluded from the nucleus. Together, these findings suggest that the primary function of the multisynthetase complex is not to aid bulk translation but instead to control localization of aminoacyl-tRNA synthetases in the cell and possibly sequester them until release upon a cue.
While our investigation into the hidden roles of the multisynthetase complex arose as a curiosity driven project, it carries implications for human health. Mutations in aminoacyl-tRNA synthetase cause white matter disorders in the brain. In a subset of patients, the mutations do not affect the enzymatic activity of the protein but instead its association with other components of the multisynthetase complex. We are currently investigating functions of aminoacyl-tRNA synthetases in the nucleus to further understand the necessity of their subcellular localization. The support through HFSP allowed us to thoroughly investigate an enigmatic complex that yielded predominantly what is traditionally thought of as negative data – at a first glance, not much changes upon the loss of two enzymes from the complex. Here, this was exactly what was so surprising and allowed us a glimpse into the hidden nature of these ancient enzymes.
Press release from Scripps Research Institute
|
HFSP award information Long-Term Fellowship (LT000207/2017-L): Elucidating Arginyl-tRNA synthetase moonlighting functions with mass spectrometry-based terminomics Fellow: Haissi Cui |


































