Human cells are constantly exposed to different cues from the environment or from changes in their internal states. Faced with these changes, cells need to make decisions and ultimately trigger responses that adapt the cell state. One of the fastest mechanisms by which cells can modulate their internal conditions is the transient phosphorylation of proteins. Catalysed by the action of kinases, protein phosphorylation can quickly alter the function of proteins via diverse mechanisms. Importantly, kinases can also regulate each other in signalling circuits, such that their combined action can result in complex behaviours (e.g. memory or oscillations). The study of these signalling circuits has been limited by our lack of capacity to measure activity changes for large numbers of kinases across conditions. In this work, Ochoa and colleagues collected 42 publically available mass spectrometry studies quantifying close to 3 million modified peptides. By integrating these data with prior knowledge of kinase target phosphosites, the activity changes for over 200 human kinases were inferred for a broad panel of nearly 400 perturbations.
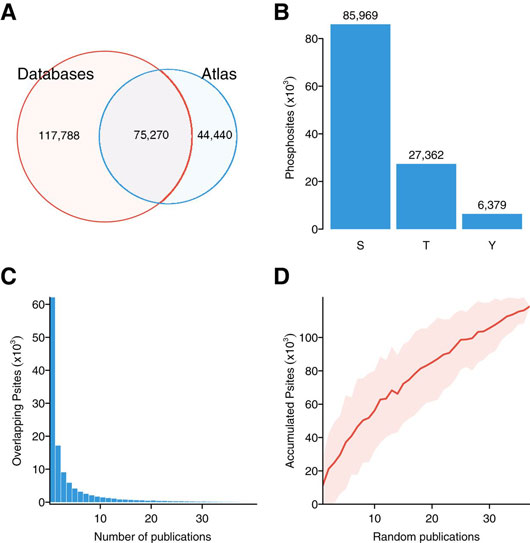
Figure: A map of human conditional phospho‐regulation. (1) Venn diagram including the total number of phosphorylated residues for which quantifications were collected and the number of sites contained in the curated phosphosite databases: PhosphoSitePlus, HPRD, and Phospho.ELM (August 2014). (2)Total number of quantified serines (S), threonines (T), and tyrosines (Y). (3) Distribution of sites reported by multiple parallel publications. (4) Accumulation of sites as publications are randomly aggregated (100 permutations). Red line shows mean and the shadowed area mean ± 1 standard deviation. Figure first appeared in Mol Syst Biol 12, 888 (2016). 10.15252/msb.20167295, Ochoa et al., An atlas of human kinase regulation.
The resulting profile of kinase regulation across conditions provides a first look at the large diversity of cell signalling states. It reveals how different perturbations present distinctive patterns of kinase activation/inhibition. For instance, the infection of human cells by two different gram-negative bacteria provokes a very similar profile of kinase activity changes. In the atlas, opposite profiles of kinase regulation were identified for cells treated with specific drugs and these drugs were then shown to influence the infection process. This illustrates how this atlas can be used to identify perturbations that may block or promote specific signalling states and/or transitions. In a different application of the kinase activity inference, the co-regulation profile of kinase activities and phosphosites was found to be predictive of kinase-kinase, kinase-sites and kinase-complexes interactions. For instance, the co-regulation of Akt kinase activity and phosphosites found in the atlas identified novel Akt target substrates.
To take full advantage of this research, a web interface is available at phosfate.com where phosphorylation signalling data can be analysed and compared with the atlas. Given the increasing amount of biological measurements available, the principles described in this research will assist others to interpret the signalling consequences in their conditions of interest and derive a much more informative diagnostic of the cellular state.
Reference
An atlas of human kinase regulation. Ochoa, D. Jonikas, M. Lawrence, R. El Debs, B. Selkrig, J. Typas, A. Villén, J. Santos, S. Beltrao, P. Mol Syst Biol 12, 888 (2016). 10.15252/msb.20167295.


































