Decades of research by the neuroscience community have revealed a great deal about the composition and structure of synapses, but how synapse formation is regulated at the gene expression level remains elusive. What are the molecular programs that instruct synapse formation so that neurons are poised to generate synaptic proteins to ensure functional connectivity?
During synaptogenesis, neurons utilize conserved activity-regulated transcriptional mechanisms to modulate expression of synaptic proteins. In vertebrate systems, the activity-dependent transcription factor Fos is critical for synapse formation and plasticity, but how Fos controls genetic pathways to modulate synaptic gene expression is unclear.
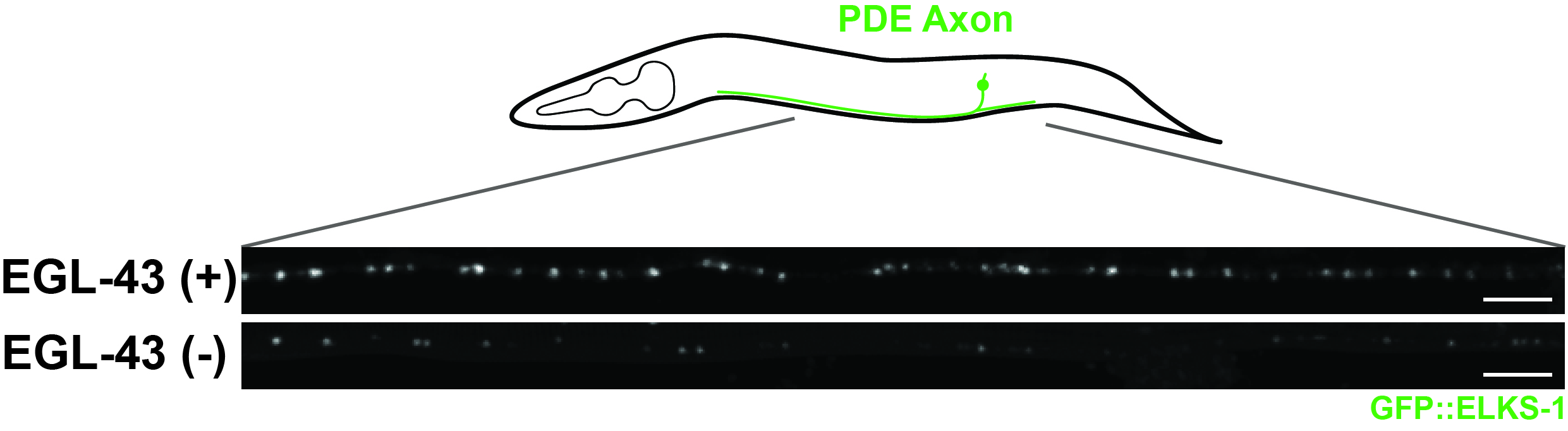
In their quest to understand how neuronal activity controls synapse formation, Stanford University scientists turned to the unique advantages of the model organism Caenorhabditis elegans (C. elegans). This organism's compact nervous system (300 neurons) and transparent nature allow for direct visualization of endogenous fluorescently labeled synaptic proteins in live animals.
Using C. elegans PDE (PosteriorDeirid) dopaminergic neurons, the researcher found that neuronal activity stimulates genetic pathways to fine-tune synaptic gene expression during development. Yee identified a conserved transcription factor, EGL-43/MECOM, that controls the expression of a network of developmental transcription factors to drive synaptogenesis in PDE. Loss of EGL-43 during development dramatically reduces the expression of synaptic proteins, such as the active zone protein ELKS-1/ELKS (Figure 1), leading to defects in a PDE-dependent behavior.
In a surprising turn, Callista Yee discovered that EGL-43 functions together in a positive feedback loop with FOS-1/FOS. Since FOS-1 expression is induced by activity, this mechanism thus enables activity to modulate the expression of genetic pathways that control synaptic gene expression. EGL-43 is expressed predominantly in sensory neurons, and the HFSP Fellowship Awardee hypothesizes that this discovered activity-regulated mechanism evolved to enable plasticity and fine-tune behavior for survival. This discovery significantly expands our understanding of synaptic protein regulation.


































