The ribosome, a fundamental component of cellular protein synthesis machinery, is one of the largest macromolecules in cells. The Escherichia coli ribosome consists of two subunits (the large and small subunit). The large subunit is assembled from 2 ribosomal RNAs (5S and 23S rRNA) and 33 ribosomal proteins, whereas the small subunit is assembled from single rRNA (16S rRNA) and 21 ribosomal proteins. In total, 3 rRNAs and 54 proteins are required to be assembled into a single macromolecular complex. Since the translation system is at the heart of self-replication, in that the same number of cellular proteins must be produced by the ribosome, studying de novo ribosome biogenesis, including its self-organization, is one of the essential factors for understanding life’s ability to self-replicate.
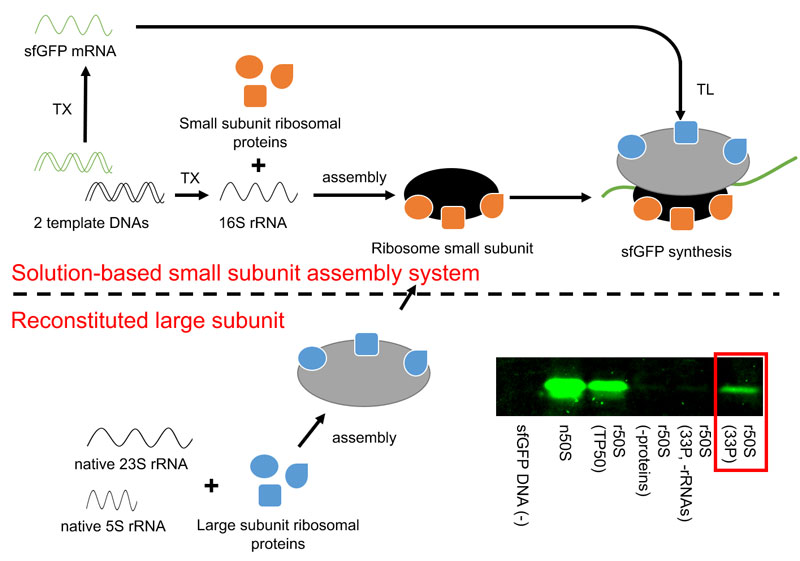
Figure: The functional large subunit assembly was achieved using native 23S and 5S rRNAs and individually prepared 33 ribosomal proteins (left side at the bottom). The assembled large subunit was further added into the previously developed solution-based small subunit assembly system (top). In this system, the small subunit is assembled from in vitro transcribed 16S rRNA and individually prepared 21 ribosomal proteins in a cell-free protein synthesis system. The function of the assembled ribosome from 54 individually prepared proteins and 3 rRNAs was addressed by superfolder green fluorescent protein synthesis in the cell-free system (the red box is the result of polyacrylamide gel electrophoresis analysis, placed at the bottom right side). The term n50S represents the native large subunit, r50S represents the reconstituted large subunit, TP50 represents native large subunit ribosomal proteins obtained from the native large subunit, and 33P represents individually prepared large subunit ribosomal proteins.
The team supported by an HFSP Program Grant (RGP0043/2017) aimed to develop de novo the synthesis and assembly system of the ribosome in a cell-free protein synthesis system. The team has focused on the reconstitution of the small subunit assembly and has developed solution-based (1) and chip-based (2) assembly systems, which are both performed in the cell-free system. The solution-based system demonstrated the functional small subunit assembly from in vitro transcribed 16S rRNA and 21 individually or partially prepared cell-free synthesized ribosomal proteins. The chip-based system enabled real-time observation of the small subunit assembly from in vitro transcribed 16S rRNA and 21 in vitro synthesized ribosomal proteins under microscopic imaging.
The research was extended to the large subunit in the present study. Protocols for individually preparing the 33 ribosomal proteins were established and the functional large subunit formation from 5S and 23S rRNA and 33 ribosomal proteins was demonstrated. Superfolder green fluorescent protein (sfGFP) was shown to be successfully synthesized by the assembled large subunit in the cell-free system. The assembled large subunit was further integrated with the solution-based small subunit assembly system. The resultant cell-free system contained all 54 recombinantly prepared ribosomal proteins and the active ribosome formation was shown by the sfGFP synthesis in the cell-free system.
As demonstrated in the small subunit study, the use of individually prepared recombinant ribosomal proteins can address the effect of compositions of ribosomal proteins on the assembly processes and functions of the subunits. For example, the function of the subunits which lack each ribosomal protein was previously examined in the solution-based system (1). Such a development will provide further insight into the nature of ribosomal self-assembly and extend our knowledge of the ribosome functions and structures. Furthermore, introduction of mutations into the recombinant ribosomal proteins will allow us to prepare a variety of ribosomes in vitro, making it easier to test mutations beneficial to the protein synthesis machinery. Thus, the present study will contribute to the future of ribosome engineering studies.
|
HFSP award information Research Grant - Program (RGP0043/2017): A PURE-ly synthetic ribosome biogenesis in DNA compartments on a chip Principal investigator: Yoshihiro Shimizu, RIKEN Center for Biosystems Dynamics Research (BDR), Osaka, Japan |


































